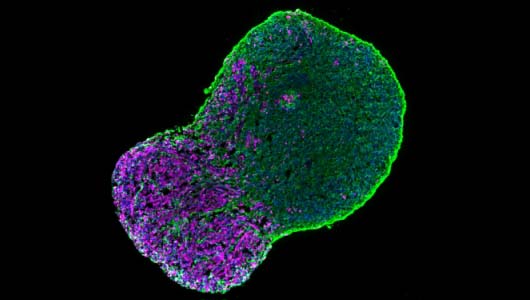
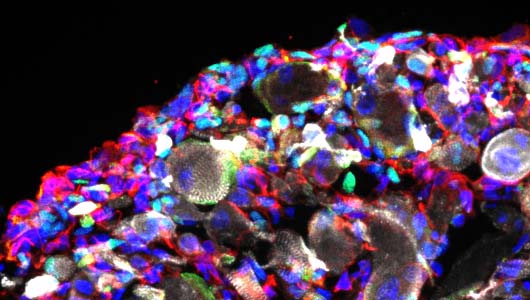
Motoneuron diseases
Our team focuses on the study and treatment of diseases affecting the communication between motor neurons and their muscle target. Using human pluripotent stem cells carrying causative mutations, we are developing new models for various neuromuscular diseases, such as Myotonia Dystrophy Type 1 or Infantile Spinal Muscular Atrophy.
Our team aims to better understand the molecular and cellular mechanisms involved in the development of diseases affecting the motor neuron and their communication with the muscle target and consequently identify new therapeutic avenues. Our research axes are declined around :
1. The generation of new models from human pluripotent stem cells to better understand the communication between motor neurons and their muscle targets. In recent years our group has developed several protocols to efficiently convert human pluripotent stem cells into different types of motor neurons. We have also established several protocols for co-culturing with muscle cells to functionally measure the communication between these two cell types.
We are now working on the establishment of 3D organoids to generate more complex models of the neuromuscular junction.
2. Deepening our knowledge of the physiopathological mechanisms involved in certain neuromuscular diseases of genetic origin. Our group is mainly interested in Dystrophic Myotonia Type 1, infantile spinal muscular atrophy and amyotrophic lateral sclerosis.
For this mechanistic research, we are interested in identifying mechanisms that may explain the degeneration or malfunction of these intercellular structures. We are also interested in understanding why certain groups of motor neurons appear to be more susceptible to certain pathological processes. Our final goal is to identify cellular abnormalities that will allow us to identify new therapeutic leads.
3. Our third research axis aims at identifying new therapeutic leads, mainly through pharmacological approaches. Our approaches are essentially based on High Content Screening techniques associated with Machine Learning.
Wolfram’s syndrome
Team members
Cécile Martinat
Team leader and Director of the INSERM unit (U861)
Sandrine Baghdoyan
Research Engineer (Inserm)
Member of the Motoneurone team since its creation, Sandrine studies the in vitro therapeutic potential of repositioned pharmacological compounds as candidates for DM1.
Judith Lorant
Associate Professor (UEVE)
Judith joined the Motoneurone team in 2021 to study the potential of extracellular vesicles as a therapeutic tool for various neuromuscular diseases.
Azania Abatan
Doctoral student
Integrated in the Motoneurone team as part of her M2 internship in February 2021, then as a study engineer in October 2021, Azania started her PhD in January 2023. She is working on the development and characterization of DM1 cortical organoids.
Morgan Gazzola
Post Doctorate (Inserm)
Joining the Motoneuron team in June 2022, Morgan is investigating the implications of certain splicing defects in the establishment of the neuromuscular junction in DM1 and the development of novel cellular models for neuromuscular diseases.
Noémie Bérenger-Currias
Research Engineer (Inserm)
Noémie joined the Motoneurones team in May 2022. She works on the TREAD screening project, in collaboration with our partners Ksilink and LDC. In particular, she is characterizing the in vitro therapeutic potential of candidate pharmacological compounds for DM1.
Laurine Merriadec
Research Assistant (CECS)
Laurine joined the Motoneurons team in March 2023. She is working on the TREAD screening project, in collaboration with our partners Ksilink and LDC, in the characterization of the in vitro therapeutic potential of candidate pharmacological compounds for DM1.
Michel Cailleret
Engineer (Inserm)
Member of I-Stem from the beginning, Michel is a specialist in genetic engineering and the generation of mutated cell lines. He is involved in many projects within the institute.
Laetitia Aubry
Associate Professor (UEVE)
Leader of the Wolfram syndrome research project, Laetitia joined the Motoneurone team in 2022.
Sandra Pourtoy
Research Assistant (CECS)
Sandra arrived at I-Stem in 2015 and works on the Wolfram syndrome research project and was attached to the Motoneurons team in 2022.
Lise Morizur
Research associate (CECS)
Specialized in the development of 3D cellular models mimicking biological processes, Lise’s interest lies in the use of 3D muscles in vitro to study myopathies.
Hugo Lenoir
Doctoral student (UMR861)
Hugo completed his M1 and M2 internships in the Motoneuron team in 2022 and 2023. He started his PhD in October 2023, working on the therapeutic potential of extracellular vesicles in Duchenne muscular dystrophy.
Caroline Hermitte
Doctoral student (UMR861)
Caroline joined the motoneuron team for her Master 2 internship in January 2023, working on splicing defects involved in neuromuscular junction establishment in Dystrophic Myotonia Type 1 (DM1). Since November 2023, she has been a PhD student at I-Stem, working on the development of neuromuscular organoids in the pathologies of Dystrophic Myotonia Type 1 and infantile spinal muscular atrophy (SMA).
Collaborations
ANR PRCE : Collaboration to identify new pharmacological leads for Steinert’s Myotonia.
ANR PRC : This collaboration aims to better understand the physiopathological mechanisms involved in the development of amyotrophic lateral sclerosis.
ANR PRC : This collaboration aims to better understand the pathophysiological mechanisms involved in the neurological disorders associated with Dystrophic Myotonia Type 1.
ANR PRC : This collaboration aims to better understand the consequences of alternative splicing defects associated with Dystrophic Myotonia type 1.
SMA Europe : This collaboration aims to identify new therapeutic avenues for infantile spinal muscular atrophy.
eRARE ReCognition : This collaboration aims to understand the molecular mechanisms that may be involved in the beneficial response observée dans l’essai Optimistic sur la Myotonie Dystrophique de type 1. Ce projet européen implique une dizaine d’équipes européennes et du Canada.
Publications
Mono- and Biallelic Inactivation of Huntingtin Gene in Patient-Specific Induced Pluripotent Stem Cells Reveal HTT Roles in Striatal Development and Neuronal Functions.
01 January 2024
Journal of Huntington's disease
Nucleolar reorganization after cellular stress is orchestrated by SMN shuttling between nuclear compartments.
15 November 2023
Nature communications
Molecular Analysis of a Congenital Myasthenic Syndrome Due to a Pathogenic Variant Affecting the C-Terminus of ColQ.
11 November 2023
International journal of molecular sciences